by Shea Robison (@EpigeneticsGuy)
(Thanks for stopping by and reading this post. I’ve linked to a number of other resources in this post, so check those out as well. When you’re done, leave a comment and let me—and the others who read this post—know what you think. This post is attracting a lot of attention from around the world the past couple of days, so this post is a good opportunity to interact with others interested in genomic and epigenetics. Check out more of my posts on epigenetics through the menu at the top of this page and the lists on the righthand margin.)
by Shea Robison
A recent tweet from @dgmacarthur recommended an article from The Telegraph (UK) newspaper about “an investment of almost £300 million in Genomics England, a new initiative to read and decipher the genetic codes of 100,000 NHS patients.”
The author of this article supports this investment in genomics, but paradoxically opens the article with a personal account of the wildly disparate results of three different genetic tests he submitted a few years ago. He writes that “though each service examined the same DNA, and evaluated the same diseases, each set of results was dramatically different. My lifetime risk of glaucoma, an eye condition, was as low as 2 per cent or as high as 36 per cent, depending on which service I chose to believe. One company thought my chances of a heart attack were twice as high as another,” and that his experience was the sort of experience that “has fed a growing sense that the sequencing of the human genome…as the future of medicine, has failed to deliver on its promise.”
However, despite these disappointments, the balance of the article is about the practical benefits of genomics which justify such a significant investment. The author concedes that while genomics has not been successful in unlocking the genetic factors for most common diseases, “quite staggering progress” has been made for cancers, rare congenital diseases and infections. In particular, the author notes, genomics is benefiting those already sick – those who have certain kinds of tumor,families with congenital disorders, and the tracking of infectious diseases. According to the author, genomics does not yet allow for much prediction of illness or disease which could benefit the “worried well,” but its benefits in these other areas already justify further investment.
I don’t have any issue with anything the author has written; I assume he is correct about the shortcomings and benefits of genomics. My question, and how this relates to epigenetics, is ‘the dog that doesn’t bark’ – or what the author doesn’t mention.
As discussed in significant detail in this article, genomics relies on genome-wide association studies (GWASs) and genome-wide complex trait analyses (GCTAs) to identify the genetic components of traits and diseases. One of the primary problems with these efforts to locate the cause of diseases and really almost any traits in the genes is the complex nature of interactions between genes and between genes and the environment – encapsulated by what is called the “missing heritability problem” (e.g., traits like intelligence have been found to be highly heritable, but why have no genes for intelligence been identified? If there is such a strong hereditary component to such traits, it seems reasonable to assume identifying the genetic basis for these traits would be relatively straightforward).
GWASs and GCTAs may be able to identify specific genes and sequences of genes which are correlated with specific outcomes, but this seems like only one part of the equation. The problem with such approaches, the author observes is that “the simplistic model of additive genetic variance upon which GCTA relies (and which assumes no epistasis and no gene x environment interactions) is out of touch with current understanding of the complex, multifactorial nature of most human traits that have a genetic component.” This is where epigenetics obviously enters the picture.
In this article, though, the author refers to an aspect of genetics that is new to me as a confounding factor for GWASs and GCTAs, writing that “there is now overwhelming scientific evidence…that persons do not possess a single NUCLEAR genome [and] that the normal human condition is one of SOMATIC MOSAICISM.” Somatic mosaicism is when different cells and tissues contain different DNA sequences (or ‘genomes’), and appears to be particularly prevalent in the human brain. To the degree somatic mosaicism is legitimate, the results of a GWAS or GCTA from one part of the body will be different from a GWAS or GCTA from another part, and may not be generalizable even from within the same person. This is the first I have heard of such a thing, and I would have to investigate it much further, but the implications for genomics and for the assumptions of orthodox genetics of this somatic mosaicism are quite obvious.
Even so, this is not what initially prompted my interest about the implications for epigenetics from the article in the Telegraph: GWASs and GCTAs can identify sequences of genes, but this sequence is only one part of how genes function; another critical aspect of gene function pertains to its structure, and the role of chromatin and histones.
Chromatin are the end product of DNA being tightly wrapped around proteins called histones. These chromatin are then also tightly wrapped together which bundling allows the meters of DNA to be packed into cell nuclei only 5 to 10um in diameter. This tight bundling gives DNA its unique three-dimensional structure which also has a significant effect on genetic expression.
According to conventional genetics, what is determined by natural selection is the sequence of genes in DNA and their expression, both of which are for all practical purposes immutable; in other words, the sequences of genes in a tightly-packed segment of DNA are fixed, and then switched on or off in specific patterns to produce ostensibly more adaptive patterns of gene expression.
However, according to the research in epigenetics, sections of DNA can be acetylated causing the chromatin to relax and open up, exposing different gene sequences to methylation which can then change the pattern of on/off switches and the nature of the gene expression; this chromatin relaxation also changes the relative proximity of different genes and gene sequences which can also affect the pattern of expression – all without changing the actual DNA sequence in any way.[1] Chromatin remodeling is an important step in cell growth and differentiation, and thus modifications of chromatin remodeling functions are associated with a number of cell-based diseases such as cancer,[2] and are also now associated with obesity.[3]
Those genes which are located on the outside of a bundle are the most accessible to processes like methylation, and thus are most susceptible to changes in suppression or expression; vice versa, those genes located inside a bundle are likewise shielded from changes in expression. Also, the three-dimensional structure produced by the wrapping of chromatin places some genes adjacent to each other which are actually quite distant sequentially, which proximity also impacts the expression of these gene loci in ways which are not detectable using conventional gene sequencing.
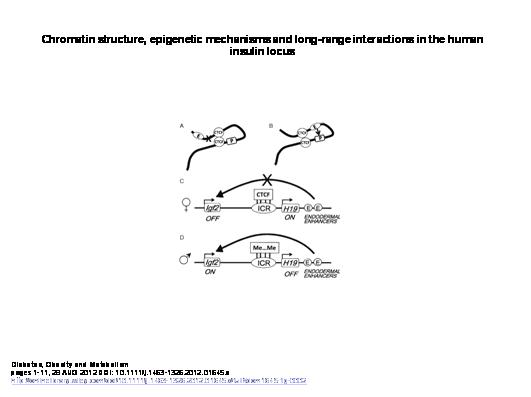
A specific example of the effects of gene structure on expression is discussed in this paper[4], which establishes “the existence of a regulatory network between the INS [human insulin] gene and other distant genes through long-range physical interactions, and suggest that such networks may have general importance for insulin biology and diabetes.” As the authors of this study observe, “of necessity, chromatin is not linear in the nucleus, but compacted to fit into the small nuclear volume. However, packing is not random; rather, it is dynamically arranged into functional loops,” and that “these chromatin loops are usually established in a cell type-specific manner, through the formation of long-range chromatin interactions mediated by proteins bound at specific DNA regulatory elements (e.g. enhancers, promoters, insulators).” As shown in the figure below, depending on the structure of a loop these enhancers and promoters can either be in positions to interact or insulated from each other:
In other words, genes can be distant from each other in sequence, which distance seems to logically preclude any significant interaction; however, owing to the physical structure of DNA, genes which are sequentially distant can actually be close to each other in three-dimensional space, which then allows them to interact in ways which are not detectable using conventional sequencing. And this structure is determined and manipulable through epigenetic mechanisms.
This conclusion returns me to the original Telegraph article and the significant investment in genomics and its standard gene-sequencing methods: If both sequence and structure play such important roles in gene expression, it seems only reasonable that considerations of both be included in any attempts to diagnose or predict genetic effects; therefore, funding for epigenetics should likewise be included on a par with funding for genomics – otherwise, we are only getting at most half of the story (though more likely genomics gives at best a third of the story, as it excludes gene structure and environmental interactions via the epigenome).
It may be that genomics already accounts for these considerations of three-dimensional structure and chromatin remodeling and so on. I don’t know; I am a political scientist with a professional interest in genetics and an enthusiastic dilettante’s understanding. If these considerations are already accounted for within genomics, and accounted for properly, I would welcome some well-intentioned enlightenment from those who know better than I.
However, what I have observed from my outsider’s perspective is that many genomicists seem remarkably antagonistic towards epigenetics. Apart from legitimate questions about the scientific validity of many of the results from epigenetics, this unwarranted antagonism often seems a function of the competition for scarce funding, or overzealous turf defending, or any number of other non-science-related factors (some of which are discussed here and here). A better approach, as discussed here by Randy Jirtle, is to promote the integration of epigenetics and genomics. This way as little as possible is left out of the equation. Otherwise, I’m not sure what we end up with.
I am curious to hear what you think. Leave your comments below and I will respond.
Also, if you find these thoughts I’ve shared interesting and worthwhile, Like this post, Reblog it, or Tweet about it using the buttons below.
[1] Thurman, R., Rynes, E. and Humbert, R. (2012). The accessible chromatin landscape of the human genome. Nature 489(7414), 75-82.
[2] Crews, D., & McLachlan, J. (2006). Epigenetics, Evolution, Endocrine Disruption, Health, and Disease. Endocrinology 147(6), s4-s10; Lehner, B.(2012). Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488, 504-507.
[3] Wang, F., Liu, H., Blanton, W., Belkina, A., Lebrasseur, N., & Denis, G. (2010). Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem. J, 425, 71-83.
[4] Xu, Z., Lefevere, G., & Felsenfeld, G. (2012). Chromatin structure, epigenetic mechanisms and long-range interactions in the human insulin locus. Diabetes, Obesity and Metabolism 14(s3), 1-11.

I couldn’t agree more on some of these points. Research needs to focus on the complete picture, which is how genetic variation reacts with the epigenome to produce our phenotypes. Most just look at the genetic contribution and ignore the epigenome, even though it can explain that missing heritability that is written off by many as “environment”, which leads them to another roll of the dice. Epigenetics isn’t a separate field but rather the next evolutionary phase of Genetics. The disciplines are so intimately intertwined that they shouldn’t be considered separate disciplines. I really hope GWAS evolves, but then again, that will lead to some pretty scary findings that most won’t want to hear. I also fear that this type of research instills fear in a lot of researchers on a career path that want to avoid controversy due to a lack of public communication about this emerging field. This emerging science needs more public resources like this and Randy Jirtle’s outreach. Keep up the great work!
LikeLike